The Context: The world, after a throbbing pandemic, is under the grasp of yet another zoonotic disaster called MonkeyPox. A recent study revealed that the rate of genetic changes in the monkeypox virus was higher than expected. In this article, we will analyse the ills of monkeypox and possible solutions from the UPSC perspective.
ABOUT THE MONKEYPOX VIRUS
MONKEYPOX
- Monkeypox is a viral zoonosis (a virus transmitted to humans from animals) with symptoms similar to those
 seen in the past in smallpox patients, although it is clinically less severe.
seen in the past in smallpox patients, although it is clinically less severe. - With the eradication of smallpox in 1980 and the subsequent cessation of smallpox vaccination, monkeypox has emerged as the most important orthopoxvirus for public health.
- Monkeypox primarily occurs in central and west Africa, often in proximity to tropical rainforests, and has been increasingly appearing in urban areas. Animal hosts include a range of rodents and non-human primates.
- Ever since it was first reported in humans in 1970, monkeypox virus infections have been largely restricted to countries in Central and Eastern Africa until recently.
RECURRENCE OF MONKEYPOX
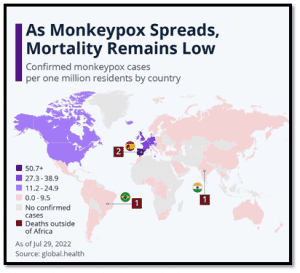
- Early in 2022, multiple cases were identified in Spain and several cases were reported from countries where the disease is not endemic, including regions in Europe and North America, and in patients with no history of travel to endemic regions.
- Following a rapid rise in cases, the World Health Organization (WHO), on July 23, 2022, declared the 2022 monkeypox outbreak as a Public Health Emergency of International Concern (PHEIC).
- As of early August 2022, over 25,000 cases of monkeypox have been reported from 83 countries, 76 of which have never historically reported monkeypox.
VIRUSES: A BASIC STUDY
DEFINITION
- A virus is an infectious microbe consisting of a segment of nucleic acid (either DNA or RNA) surrounded by a protein coat.
- Viruses are non-cellular organisms that are characterized by having an inert crystalline structure outside the living cell.
WORKING & FEATURES
- A virus cannot replicate alone; instead, it must infect cells and use components of the host cell to make copies of itself.
- Often, a virus ends up killing the host cell in the process, causing damage to the host organism. Well-known examples of viruses causing human disease include AIDS, COVID-19, measles and smallpox.
- In addition to proteins, viruses also contain genetic material, which could be either RNA or DNA.
- No virus contains both RNA & DNA.
- Viruses that infect plants have single-stranded RNA & Viruses that infect animals have either single or double-stranded RNA or double-stranded DNA.
- Bacteriophages (viruses that infect bacteria) are usually double-stranded DNA viruses.
TYPES
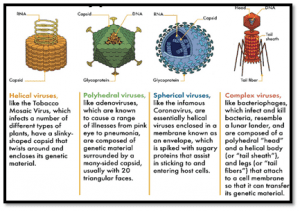
DIFFERENCES BETWEEN DNA AND RNA VIRUSES
- DNA viruses contain DNA as the genetic material while RNA viruses contain RNA as the genetic material. Some examples of DNA viruses are Herpes viruses, poxviruses, and hepatitis B.
- Generally, DNA genomes are larger than RNA genomes. Furthermore, most DNA viruses contain double-stranded DNA while most RNA viruses contain single-stranded RNA. Rhabdovirus, coronavirus, SARS, poliovirus, rhinovirus, hepatitis A virus, influenza virus, etc., are some examples of RNA viruses.
PROLIFERATING GENETIC MUTATIONS: THE EVOLUTION AND CONCERN
Recently, a team of researchers at the National Institute of Health Doutor Ricardo Jorge in Portugal has found that the monkeypox virus has been evolving at a faster rate than expected.
KEY FINDINGS
- Scientists said the latest strain of monkeypox, once previously confined to parts of Africa, has about 50 genetic variations compared to related viruses that circulated in 2018-2019.
- They found the virus is continuing to evolve during the current outbreak, including a number of small changes in the genetic code, minor gene variants and a deleted gene.
LOOMING CONCERNS
- The monkeypox virus has a DNA genome of around 2,00,000 base pairs, roughly six times larger than that of SARS-CoV-2. Like other viruses, the monkeypox virus evolves by the accumulation of genetic errors, or mutations, in its genome when it replicates inside a host.
- Information about mutations occurring in different genome sequences of the monkeypox virus across different regions can, thus, provide essential insights into how the virus is evolving, its genetic diversity and other factors that may be relevant to the development of diagnostic tools.
RISKS OF PARALLEL EVOLUTION
- Being a DNA virus, the monkeypox virus, like other poxviruses, was believed to have a small rate of accumulating genetic changes compared to viruses with an RNA genome like SARS-CoV-2, which have a much larger rate of mutations. For poxviruses, this rate is estimated to be as low as a couple of genetic changes every year.
- A recent study, however, revealed that the observed rate of genetic changes in the virus was higher than the expected average of around 50 genetic changes.
- The higher-than-expected rate of evolution coupled with the rapid rise in monkeypox cases across the world could potentially be due to highly parallel evolution in a large number of individuals simultaneously, as the present outbreak came out of a super spreader event.
ASPECT OF APOBEC3 PROTEIN
- The researchers also suggest that several mutations that have been identified in the new sequences of the monkeypox virus may have emerged due to interaction between the virus genome and an important family of proteins coded by the human genome known as the Apolipoprotein B Editing Complex (or APOBEC3). These proteins offer protection against certain viral infections by editing the genome sequence of the virus while it replicates in the cell.
- Therefore, researchers suggest that many of the genetic mutations in the monkeypox genomes from the current outbreak are remnants of the effect of APOBEC3 and may not provide a significant evolutionary advantage to the virus.
POSSIBLE OUTREACH OF THE VIRUS
- Monkeypox virus can infect a range of hosts, including non-human primates and rodents, which could act as a natural reservoir. Infections in the reservoir could also enable continued transmission and accumulation of mutations before spilling over to cause human infections.
MONKEYPOX LINEAGES
- Clusters of genomes having common and shared mutations and a common origin are referred to as a lineage or clade. In the early 2000s, two different clades of monkeypox virus were defined in Africa, where several cases of the disease have been seen the Central African (Congo Basin) clade and the West African clade, of which the Congo Basin clade has been shown to be more transmissible and cause more severe disease.
HOW ARE LINEAGES OF VIRUSES NAMED?
- Since naming viral lineages using the country or geography of origin could be discriminatory and possibly not in the right spirit, a new system of naming monkeypox lineages has been proposed by researchers recently.
- Under the new proposed system, the Congo Basin clade is denoted as clade 1, while the West African clade is divided into clade 2 and clade 3.
- This new system will also describe sub-lineages of the virus, with the original parent lineage being denoted as lineage ‘A’, and its descendants as ‘A.1’, ‘A.1.1’, ‘A.2’, and ‘B.1’.
- Lineage B.1 denoted the current 2022 outbreak of monkeypox virus infections which is a descendant of the A.1.1 lineage.
SIGNIFICANCE OF LATEST MONKEYPOX OUTBREAK
UNDERSTANDING GEOGRAPHICAL DISTRIBUTION OF THE VIRUS
- With several genome sequences of the monkeypox virus available in public databases, it is possible today to understand the prevalence of different lineages of the virus across different regions.
- Over 95% of the recently deposited genome sequences of the virus belong to the B.1 lineage of the monkeypox virus, and this lineage is epidemiologically linked to the super spreader events in Europe that formed the basis for the current outbreak of monkeypox.
TRACKING THE SPREAD
- While a majority of the genomes deposited could be linked to the 2022 outbreak of monkeypox, sequences deposited recently in 2022 from the U.S., Thailand and India suggest that there is a second distinct lineage of the monkeypox virus that is currently in active circulation in 2022.
- These genomes are classified as the A.2 lineage of the monkeypox virus and currently encompass six genome sequences, including two that were collected from Kerala.
- The earliest genome belonging to this lineage was collected from Texas in 2021, while the two sequences from Kerala collected in 2022 cluster closely with a genome collected from Florida in the same year.
AIDING GENOMIC SURVEILLANCE
- Genomic surveillance of pathogens provides interesting insights by following a molecular approach for contact tracing and understanding the transmission of the virus across the world.
- As cases of monkeypox continue to rise, it is therefore important to strengthen the genomic surveillance for the monkeypox virus.
- Since data from the present outbreak suggest a sustained human-to-human transmission, continuous genomic surveillance is important to understand the evolution and adaptation of the virus, apart from providing useful data to epidemiologists.
THE WAY FORWARD
- Genomic surveillance has played a crucial role in the global Covid -19 response, with countries like South Africa able to make essential contributions in detecting variants due to their capacities in this area. Thus, in the wake of the COVID-19 cases continuing unabated and monkeypox having a proliferating trend, there is an urgent need to build a sustainable system for genomic surveillance in India.
- Genomic sequencing is a crucial part of every country’s approach to detecting and containing outbreaks of other pathogens. Indian SARS-CoV-2 Consortium on Genomics (INSACOG) has conducted various surveillance studies on genomic mutations. It provides an accurate real-time picture of how a pandemic is moving, and thus, its capabilities must be continuously refined. The INSACOG is crucial in:
- Early detection of genomic variants of public health implications through sentinel surveillance.
- To determine the genomic variants in unusual events/trends (Vaccine breakthrough, super-spreader events, high mortality/morbidity trend areas etc.).
- To correlate the genome surveillance data with epidemiological data.
- To suggest public health actions based on the analysis of genomic and epidemiological surveillance data.
- A Rapid Response Team (RRT) must be formed in each State/UT by the Health Department. The team should comprise a clinician, a microbiologist and a member of the Medical College (preferably from the Community Medicine Department). As soon as any mutation is detected and conveyed to the State/UT, the RRT must be deployed by the State/UT to the site, where it will investigate the mutant
THE CONCLUSION: Recently, WHO’s Science Council released a report, “Accelerating access to genomics for global health”, advocating for passing on Genomic Technologies to developing countries. The report mentions that countries with established expertise must come forward in support of vulnerable developing nations for the cause of enhancing their genetic sequencing and surveillance capabilities.
QUESTIONS TO PONDER
- “With COVID-19 continuing unabated and monkeypox around the corner, the time has never been better, and the need never more acute, to build a sustainable system for genomic surveillance in India.” In the light of this statement, examine the efficacy of the recently formed Indian SARS-CoV-2 Genomics Consortium (INSACOG).
- “A recent study revealed that the rate of genetic changes in the monkeypox virus was higher than expected.” In the light of this statement, explain the potential causes of the proliferation of viruses like MonkeyPox.
- Discuss the types of viruses. How are lineages of viruses named? Explain in the light of the monkeypox virus.




